What is DNA?
DNA (deoxyribonucleic acid) is a material that contains an encoded version of all the information necessary to build and maintain an organism. DNA is a nucleic acid - a large biological molecule composed of building units called nucleotides (guanine, adenine, thymine and cytosine). Most DNA molecules consist of two biopolymer strands, coiled around each other to form a double helix.
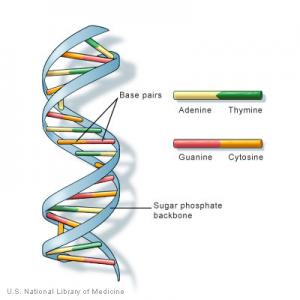
When organisms reproduce, biological information in replicated as the two strands separate and a portion of their DNA is passed along to their offspring, helping to ensure continuity between generations (while allowing changes that create diversity). A gene is a segment of DNA that is passed down from parents to children and confers a trait to the offspring. Genes are organized in units named chromosomes - humans have 23 pairs of them, one from the mother and one from the father. An entire set of genes, 46 chromosomes in total, is a genome.
All living organisms have DNA within their cells, and nearly every cell in a human being contains a full set of DNA. DNA can replicate, and each strand of the double helix can serve as a pattern for duplicating the sequence of bases from the other strand, because bases pair together in a certain way - A with T, C with G. The order of these bases determines the information for building the organism, similar to how letters of the alphabet appear in a certain order to form words.
What is DNA sequencing?
DNA sequencing is the process of determining the order of nucleotide bases in a DNA molecule. It may be used for determining sequences of specific genes, clusters of genes, full chromosomes or even entire genomes. Knowing the sequence of bases helps understand the nature of genetic information in a particular segment of DNA, which can have diverse applications like identifying gene associations with diseases, understanding developmental processes in humans, studying evolution of different species, manipulating plants and agriculture for various ends, acquiring a deeper understanding of microbes and viruses and more.

How is DNA sequencing done?
Basic DNA sequencing methods include the Maxam-Gilbert method, also known as chemical sequencing, which was developed in 1977 and is based on chemical modification of DNA and cleavage at specific bases. The methodâs use of radioactive labelling and its technical complexity hindered its extensive use. Another method invented in 1977 is the Sanger method, or chain-termination, which is relatively easy, reliable and uses fewer toxic chemicals and radioactivity. It was quickly adopted into use and further along became automated, and was even used in the first generation of DNA sequencers. Technological advances of the following decades enabled it to become more efficient and less expensive, resulting in its use in the first human genome project in 2001.
A large number of different methods and approaches reached the market following drastic technological changes, with different applications ranging from mapping an entire genome to a targeted search of specific genomic region. Some approaches are used to determine the sequence of DNA with no previously known sequence (âde novoâ methods), some are âshotgunâ sequencing, which means sequencing long strands of DNA by breaking the target DNA into random fragments and later reassembling them, and next generation methods are constantly researched, opting to lower costs, improve efficiency and cater to various sequencing needs.
Graphene and DNA sequencing
Graphene is a material made from honeycomb sheets of carbon just one atom thick. Science journals and researchers are exhausted from trying to find new superlatives for this wondrous material: it's the lightest, strongest, thinnest, best heat and electricity conducting material ever discovered. It holds promise to revolutionize everything from computing to tennis rackets.
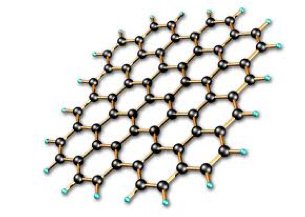
In the fields of biotechnology and medicine, it seems grapheneâs thin form and malleable nature make it well suited for many possible applications such as disease and tumor detection, drug delivery, DNA sequencing and more. In DNA sequencing, a main concept is creating a graphene membrane, immerse it in conductive fluid and apply a voltage to one end so DNA can be drawn through the grapheneâs miniscule pores. This method is called nanopore sequencing and it would allow DNA to be analyzed one nucleotide at a time (each nucleotide effecting the membrane differently due to its unique dimensions and electrical properties). Additional concepts involve graphene-based DNA sensors, and alternative ways of making DNA sequencing faster and more efficient.