Researchers from the University of Illinois at Urbana-Champaign (UIUC) have developed a new nanopore sequencing method based on graphene nanoribbons that detects double and single stranded DNA in different configurations. This graphene-based detector shows great sensitivity and holds promise for developing a portable, high-throughput sequencer that can also detect DNA morphological transformations.
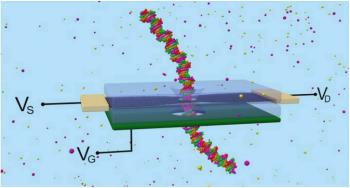
In a nanopore sequencing reaction, DNA passes through a nanopore drilled in a membrane to which an electrical voltage is applied. When DNA goes through the pore, it causes dips in the electrical current. Reading the magnitude and duration of the electrical changes allows to identify the bases that go through.
Recent advances allowed researchers to develop graphene nanoribbons (GNRs) that could sequence ssDNA and dsDNA in different configurations. The graphene layer one-atom thickness provides unmatched sequencing resolution. Additionally, graphene allows electric control of the biomolecules passing through the pore. A back-gate controls the carrier concentration in the graphene, enabling to optimize sensitivity to the nucleotides. The graphene irregular shape generates many regions of great electronic sensitivity.
The nanodevice was able to detect the helical structure of dsDNA and its helix-to-ladder forced conformational transition. Its high sensitivity, adjustable with changes in carrier concentration or nanopore geometry, allowed to sequence a ssDNA strand.