A team led by Boston College researchers has used a sheet of graphene to track the electronic signals inherent in biological structures, in order to develop a platform to selectively identify deadly strains of bacteria. This effort could lead to more accurate targeting of infections with appropriate antibiotics, according to the team.
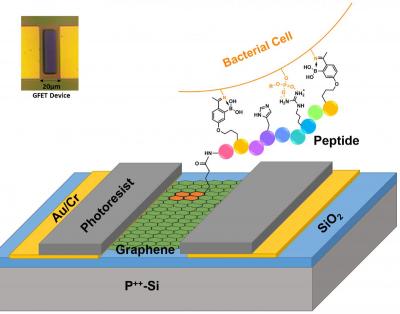
The prototype demonstrates the first selective, rapid, and inexpensive electrical detection of the pathogenic bacterial species Staphylococcus aureus and antibiotic resistant Acinetobacter baumannii on a single platform, said Boston College Professor of Physics Kenneth Burch, a lead co-author of the paper.
The rapid increase in antibiotic resistant pathogenic bacteria has become a global threat, in large part because of the over prescription of antibiotics. This is driven largely by the lack of fast, cheap, scalable, and accurate diagnostics, according to co-author and Boston College Associate Professor of Biology Tim van Opijnen.
Particularly crucial is identifying the bacterial species and whether it is resistant to antibiotics, and to do so in a platform which can be easily operated at the majority of points of care. Currently such diagnostics are relatively slow - taking from hours to days - require extensive expertise, and very expensive equipment.
The Boston College researchers, working with colleagues from Boston University, developed a sensor, known as a graphene field effect transistor (G-FET), that can overcome critical shortcomings of prior detection efforts since it is a highly scalable platform that employs peptides, chains of multiple linked amino acids, which are inexpensive and easy-to-use chemical agents, according to co-author and BC Professor of Chemistry Jianmin Gao.
The team set out to show it could construct a device that can "rapidly detect the presence of specific bacterial strains and species, exploiting the large amount of electric charge on their surface and ability to capture them with synthetic peptides of our own design," said Burch.
The initiative built upon the earlier research of van Opijnen and Gao, who previously found peptides were highly selective, but at that time required expensive fluorescence microscopes for their detection.
The team modified existing peptides to allow them to attach to graphene. The peptides were designed to bind to specific bacteria, rejecting all others. In essence, the G-FET is able to monitor the electric charge on the graphene, while exposing it to various biological agents.
Due to the selectivity of the peptides, the researchers were able to pinpoint their attachment to the desired bacterial strain. By electrically monitoring the resistance and, ultimately, charge on the device, the presence of bacteria attached to graphene could be resolved, even for just a single cell.
To enable greater speed and high sensitivity, an electrical field was placed on the liquid to drive the bacteria to the device, again exploiting the charge on the bacteria, the team reported. This process, known as dielectrocphoresis, had never previously been applied to graphene-based sensors and could potentially open the door to dramatically improving efforts in that field to employ graphene for biosensing, the team stated.
"We were surprised how well the bacteria were electrically guided to the devices," said Burch. "We thought it would somewhat reduce the required time and needed concentration. Instead, it worked so well that the electric field was able to bring needed concentration of bacteria down by a factor of 1000, and reduce the time to detection to five minutes."
The team is now working to expand the number of pathogens that can be detected and demonstrate that it can work directly with samples from patients.

