Graphene and DNA sequencing: introduction and market status
What is DNA?
DNA (deoxyribonucleic acid) is a material that contains an encoded version of all the information necessary to build and maintain an organism. DNA is a nucleic acid - a large biological molecule composed of building units called nucleotides (guanine, adenine, thymine and cytosine). Most DNA molecules consist of two biopolymer strands, coiled around each other to form a double helix.
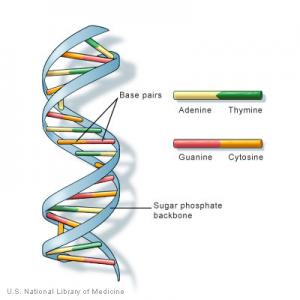
When organisms reproduce, biological information in replicated as the two strands separate and a portion of their DNA is passed along to their offspring, helping to ensure continuity between generations (while allowing changes that create diversity). A gene is a segment of DNA that is passed down from parents to children and confers a trait to the offspring. Genes are organized in units named chromosomes - humans have 23 pairs of them, one from the mother and one from the father. An entire set of genes, 46 chromosomes in total, is a genome.
All living organisms have DNA within their cells, and nearly every cell in a human being contains a full set of DNA. DNA can replicate, and each strand of the double helix can serve as a pattern for duplicating the sequence of bases from the other strand, because bases pair together in a certain way - A with T, C with G. The order of these bases determines the information for building the organism, similar to how letters of the alphabet appear in a certain order to form words.
What is DNA sequencing?
DNA sequencing is the process of determining the order of nucleotide bases in a DNA molecule. It may be used for determining sequences of specific genes, clusters of genes, full chromosomes or even entire genomes. Knowing the sequence of bases helps understand the nature of genetic information in a particular segment of DNA, which can have diverse applications like identifying gene associations with diseases, understanding developmental processes in humans, studying evolution of different species, manipulating plants and agriculture for various ends, acquiring a deeper understanding of microbes and viruses and more.

How is DNA sequencing done?
Basic DNA sequencing methods include the Maxam-Gilbert method, also known as chemical sequencing, which was developed in 1977 and is based on chemical modification of DNA and cleavage at specific bases. The method’s use of radioactive labelling and its technical complexity hindered its extensive use. Another method invented in 1977 is the Sanger method, or chain-termination, which is relatively easy, reliable and uses fewer toxic chemicals and radioactivity. It was quickly adopted into use and further along became automated, and was even used in the first generation of DNA sequencers. Technological advances of the following decades enabled it to become more efficient and less expensive, resulting in its use in the first human genome project in 2001.
A large number of different methods and approaches reached the market following drastic technological changes, with different applications ranging from mapping an entire genome to a targeted search of specific genomic region. Some approaches are used to determine the sequence of DNA with no previously known sequence (“de novo†methods), some are “shotgun†sequencing, which means sequencing long strands of DNA by breaking the target DNA into random fragments and later reassembling them, and next generation methods are constantly researched, opting to lower costs, improve efficiency and cater to various sequencing needs.
Graphene and DNA sequencing
Graphene is a material made from honeycomb sheets of carbon just one atom thick. Science journals and researchers are exhausted from trying to find new superlatives for this wondrous material: it's the lightest, strongest, thinnest, best heat and electricity conducting material ever discovered. It holds promise to revolutionize everything from computing to tennis rackets.
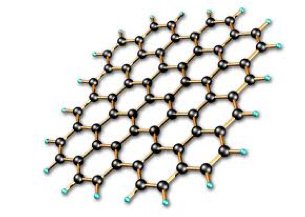
In the fields of biotechnology and medicine, it seems graphene’s thin form and malleable nature make it well suited for many possible applications such as disease and tumor detection, drug delivery, DNA sequencing and more. In DNA sequencing, a main concept is creating a graphene membrane, immerse it in conductive fluid and apply a voltage to one end so DNA can be drawn through the graphene’s miniscule pores. This method is called nanopore sequencing and it would allow DNA to be analyzed one nucleotide at a time (each nucleotide effecting the membrane differently due to its unique dimensions and electrical properties). Additional concepts involve graphene-based DNA sensors, and alternative ways of making DNA sequencing faster and more efficient.
Further reading
- Introduction to graphene
- Graphene company database
- How to invest in the graphene revolution
- The Graphene Handbook, our very own guide to the graphene market
Crumpled graphene could enable fast, simple and sensitive biosensors
Researchers at the University of Illinois at Urbana-Champaign have found that crumpling graphene makes it more than ten thousand times more sensitive to DNA by creating electrical "hot spots". This discovery could assist in addressing a known issue of graphene-based biosensors - the face that they require a lot of DNA in order to function properly.
"This sensor can detect ultra-low concentrations of molecules that are markers of disease, which is important for early diagnosis," said study leader Rashid Bashir, a professor of bioengineering and the dean of the Grainger College of Engineering at Illinois. "It's very sensitive, it's low-cost, it's easy to use, and it's using graphene in a new way."
Cardea Bio and Nanosens Innovations merger-acquisition finalized
Cardea Bio (formerly: Nanomedical Diagnostics) and Nanosens Innovations have joined forces to accelerate the development of the Genome Sensor: the world's first DNA search engine that runs on CRISPR-Chip technology.
Cardea has announced the finalization of their merger-acquisition of Nanosens Innovations, the creators of CRISPR-Chip. Cardea first came out with the news of the proposed merger in September, along with the announcement of their Early Access Program for the Genome Sensor. Built with CRISPR-Chip technology, the Genome Sensor is the world’s first DNA search engine. It can google genomes to detect genetic mutations and variations.
University of Illinois team finds that defects in graphene membranes may improve biomolecule transport
Researchers at the University of Illinois examined how tiny defects in graphene membranes, formed during fabrication, could be used to improve molecule transport. They found that the defects make a big difference in how molecules move along a membrane surface. Instead of trying to fix these flaws, the team set out to use them to help direct molecules into the membrane pores.
Nanopore membranes have generated interest in biomedical research because they help researchers investigate individual molecules - atom by atom - by pulling them through pores for physical and chemical characterization. This technology could ultimately lead to devices that can quickly sequence DNA, RNA or proteins.
Graphene crinkles can be used as 'molecular zippers'
Scientists have noticed many years ago that when buckyballs (soccer ball shaped carbon molecules) are thrown onto a certain type of multilayer graphene, they spontaneously assemble into single-file chains that stretched across the graphene surface. Now, researchers from Brown University have explained how the phenomenon works, and that explanation could pave the way for a new type of controlled molecular self-assembly.
 BUCKYBALLS LINED UP ON A LAYERED GRAPHENE SURFACE
BUCKYBALLS LINED UP ON A LAYERED GRAPHENE SURFACE
The Brown team shows that tiny, electrically charged crinkles in graphene sheets can interact with molecules on the surface, arranging those molecules in electric fields along the paths of the crinkles.
Team finds that an electric field applied to a tiny hole in a graphene membrane could compress water molecules
Researchers at the University of Illinois at Urbana-Champaign have developed new theories regarding the compression of water under a high-gradient electric field. They found that a high electric field applied to a tiny hole in a graphene membrane would compress the water molecules travelling through the pore by 3%. The predicted water compression may eventually prove useful in high-precision filtering of biomolecules for biomedical research.
The team commented: "This is an unexpected phenomenon, contrary to what we thought we knew about nanopore transport. It took three years to work out what it was the simulations were showing us. After exploring many potential solutions, the breakthrough came when we realized that we should not assume water is incompressible. Now that we understand what's happening in the computer simulations, we are able to reproduce this phenomenon in theoretical calculations."
Graphene forms electrically-charged crinkles when compressed
Researchers from Brown University have discovered yet another peculiar and potentially useful property of graphene, that could be useful in guiding nanoscale self-assembly or in analyzing DNA or other biomolecules.

Their new study demonstrates mathematically what happens to stacks of graphene sheets under slight lateral compressionâa gentle squeeze from their sides. Rather than forming smooth, gently sloping warps and wrinkles across the surface, the researchers found that layered graphene forms sharp, saw-tooth ridges that turn apparently have interesting electrical properties.
Graphene-based sensors to advance diagnostic genome sequencing
University of Arkansas researchers are working together, with support from the National Institutes of Health, to make that prospect of graphene-based sensors that sequence a patient's genome to predict diseases more realistic. Steve Tung, professor of mechanical engineering, and Jin-Woo Kim, professor of biological engineering, have received a grant (of approximately $400,000) from the NIH's Human Genome Research Institute to develop nanoscale technology designed to make DNA sequencing faster, cheaper and easier.
The base of the research builds on the concept of nanochannel measurement, in which individual strands of DNA pass through a tiny channel. The passage of those strands interrupts an electrical current and a sensor detects the nature of the interruption, telling scientists which nucleotide has passed through the channel.
Researchers use graphene to increase the sensitivity of diagnostic devices
Researchers at the University of Pennsylvania have used graphene to increase the sensitivity of diagnostic devices, in particular those used to monitor and treat HIV. The team combined a trick of DNA engineering which involves an engineered piece of DNA called a hairpin, with biosensors, increasing the sensitivity of the sensors by a factor of 50,000 in less than an hour.
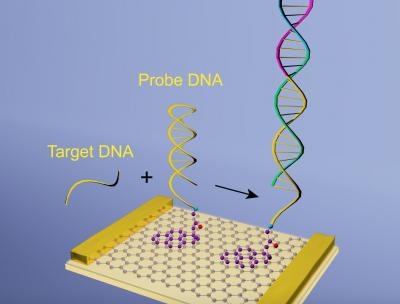
The biosensorsare made with graphene, and so can be used as an extremely sensitive way of detecting biological signals, measuring the current that flows through graphene surface in the presence of biomolecules. When DNA or RNA molecules bind to the graphene, it produces a big change in the conductivity of the atomically thin material, allowing the researchers to detect infections and to measure viral loads.
Spanish scientists open the door to using graphene in smart filters and sensors
As part of a national research collaboration, Spanish researchers including the ICN2 have reached a milestone in graphene research, that potentially brings science a step closer to using graphene in filtration and sensing applications.
The researchers have successfully synthesized a graphene membrane with pores whose size, shape and density can be tuned with atomic precision at the nanoscale. Engineering pores at the nanoscale in graphene can change its fundamental properties. It becomes permeable or sieve-like, and this change alone, combined with graphene's intrinsic strength and small dimensions, points to its future use as the most resilient, energy-efficient and selective filter for extremely small substances including greenhouse gases, salts and biomolecules.
Two graphene layers placed so close together that an electric current spontaneously jumps across could benefit DNA sequencing
Researchers at Leiden University in the Netherlands have managed to bring two graphene layers so close together that an electric current spontaneously jumps across. This could enable scientists to study the edges of graphene and use them for sequencing DNA with a precision beyond existing technologies.
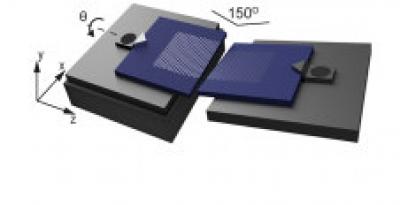
The team tilted two one-atom-thick sheets of graphene so that they only meet in one point, where electrons jump across from one layer to the other. Previous attempts with graphene electrodes failed because the layers are 'floppy' by nature. The Leiden scientists deposited them on a silicon substrate, making them rigid all the way to the edge. They brought both layers close enough together so that tunneling occursâa quantum mechanical phenomenon where electrons spontaneously jump to a neighboring material, even though there is no direct contact. Any small object in between will enhance the tunneling. The number of electrons tunneling through will tell researchers about some of its properties.
Pagination
- Page 1
- Next page

